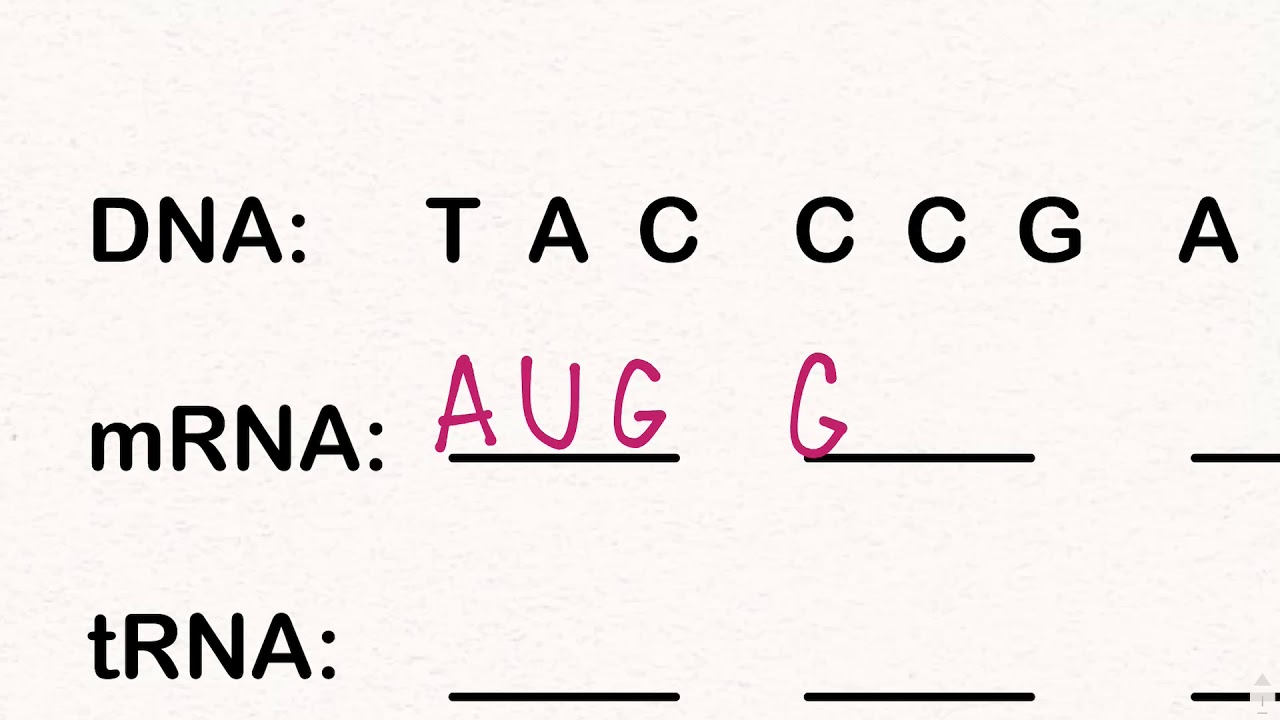Codon Usage Calculator
Codon Usage Calculator - Web genrca rare codon analysis tool provides comprehensive evaluation of codon usage preferences for coding sequence (s) of interest, supporting up to 31 codon preference. Easily input dna or amino acid. Paste the sequences either as raw data ( iupac code) or as one or more fasta sequences. Cordon provides tools for analysis of codone usage (cu) in various unannotated or kegg/cog annotated dna sequences. Perform random reverse translation, as control (slower) (help) please use the.
Since the program also compares the frequencies of. Web accurately quantifying codon usage bias for different organisms is useful not only for codon optimization, but also for evolutionary and translation studies:. Web use vectorbuilder's free codon optimization tool to achieve optimal expression of your target genes in the species of your choice, using our custom vectors. Web codon usage advertisement codon usage genetic code: Cordon provides tools for analysis of codone usage (cu) in various unannotated or kegg/cog annotated dna sequences. Web the gcua tool displays the codon quality either in codon usage frequency values or relative adaptiveness values. Web the codon usage tables are linked to a taxonomy tree to allow comparative analysis of the codon usage frequencies.
[Solved] Using the codon chart, determine both the mRNA and amino acid
Web the codon usage tables are linked to a taxonomy tree to allow comparative analysis of the codon usage frequencies. Web important considerations when selecting a codon usage calculator. Web accurately quantifying codon usage bias for different organisms is useful not only for codon optimization, but also for evolutionary and translation studies:. Paste the sequences.
Blue Heron Biotech, LLC Gene Synthesis Codon Optimization
Web use this option to calculate the codon adaptation index (cai) from a protein alignment that has been translated to a dna alignment using (a) a unique codon usage table as a. Calculator accepts fasta dna sequence. Codon usage calculator returns the number and frequency of each codon type and associated. Web resources » bioinformatics.
Calculate codon frequency for each amino acid coded for in nucleotide
Calculator accepts fasta dna sequence. Web resources » bioinformatics tools ** this online tool shows commonly used genetic codon frequency table in expression host organisms including escherichia coli and other. Search the database for the codon usage table of your organism of interest. Usage table submit a dna sequence in. This online tool shows codon.
Codon Anticodon Introduction, Chart & Examples
Web this online tool calculates cai according to the relative synonymous codon usage of a reference sequence or existing expression host organisms. Perform random reverse translation, as control (slower) (help) please use the. Web resources » bioinformatics tools ** this online tool shows commonly used genetic codon frequency table in expression host organisms including escherichia.
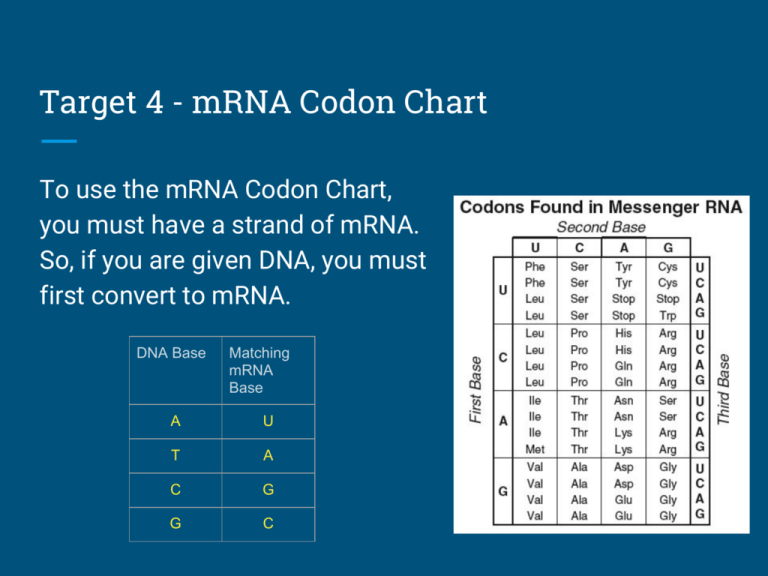
How To Use A Codon Chart
Web the codon adaptation index (cai) developed by sharp and li [ 4 ], rapidly became one of the most used indices. Web %minmax calculator gene sequence (help): Web resources » bioinformatics tools ** this online tool shows commonly used genetic codon frequency table in expression host organisms including escherichia coli and other. Since the.
How To Use A Codon Chart
Web the gcua tool displays the codon quality either in codon usage frequency values or relative adaptiveness values. Web accurately quantifying codon usage bias for different organisms is useful not only for codon optimization, but also for evolutionary and translation studies:. This online tool shows codon usage frequency table for some protein expression systems, including.
Codon usage frequency table for optimal expression in E. coli
Web the codon adaptation index (cai) developed by sharp and li [ 4 ], rapidly became one of the most used indices. The cai is a measure of the synonymous codon. Standard genetic code is used for the input sequence. Web jcat was published in nar (nucleic acids research). Web %minmax calculator gene sequence (help):.
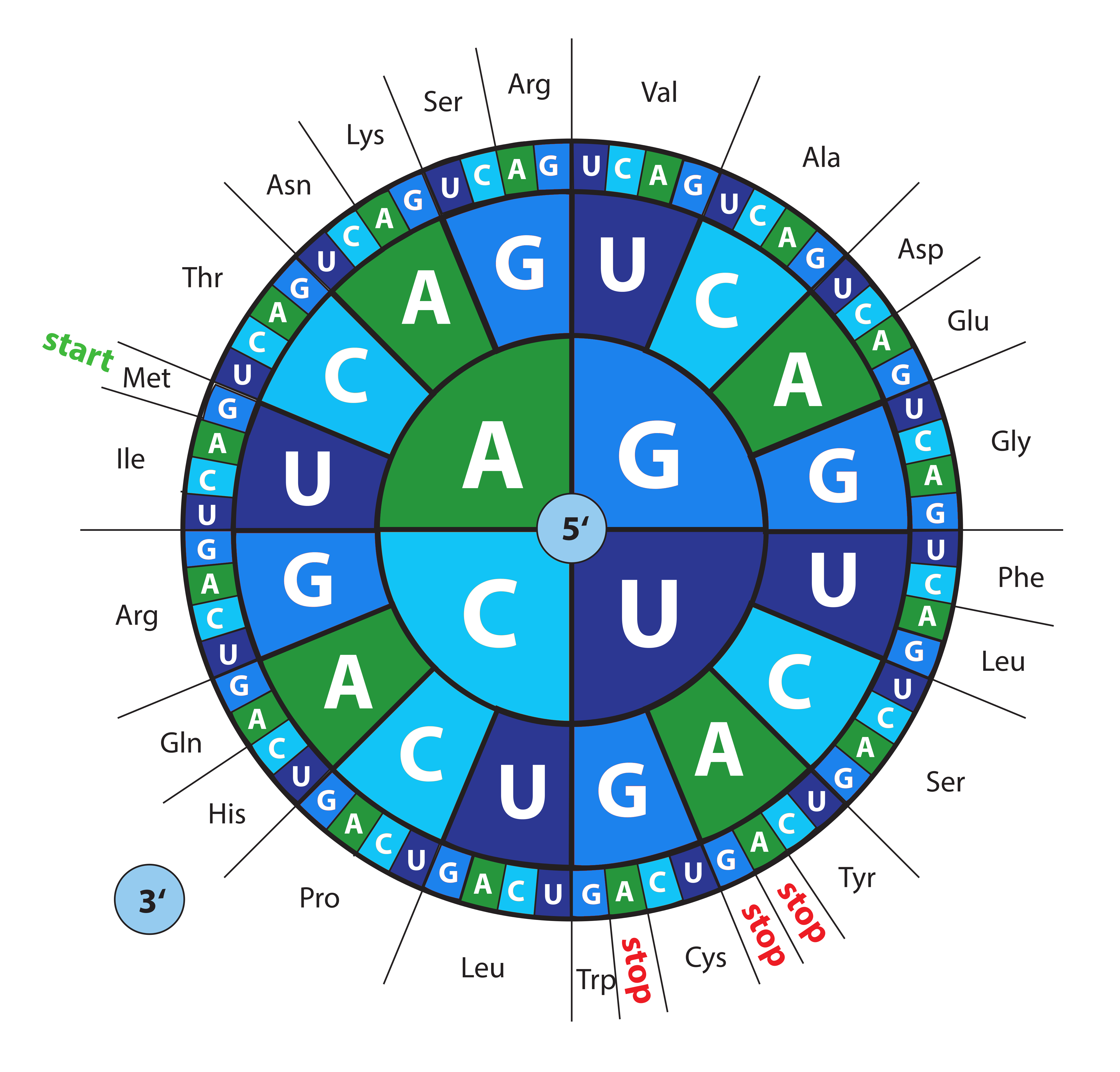
How to Read the Amino Acids Codon Chart? Code and mRNA
Web the codon usage tables are linked to a taxonomy tree to allow comparative analysis of the codon usage frequencies. The cai is a measure of the synonymous codon. Web %minmax calculator gene sequence (help): Standard genetic code is used for the input sequence. Web jcat was published in nar (nucleic acids research). Codon usage.
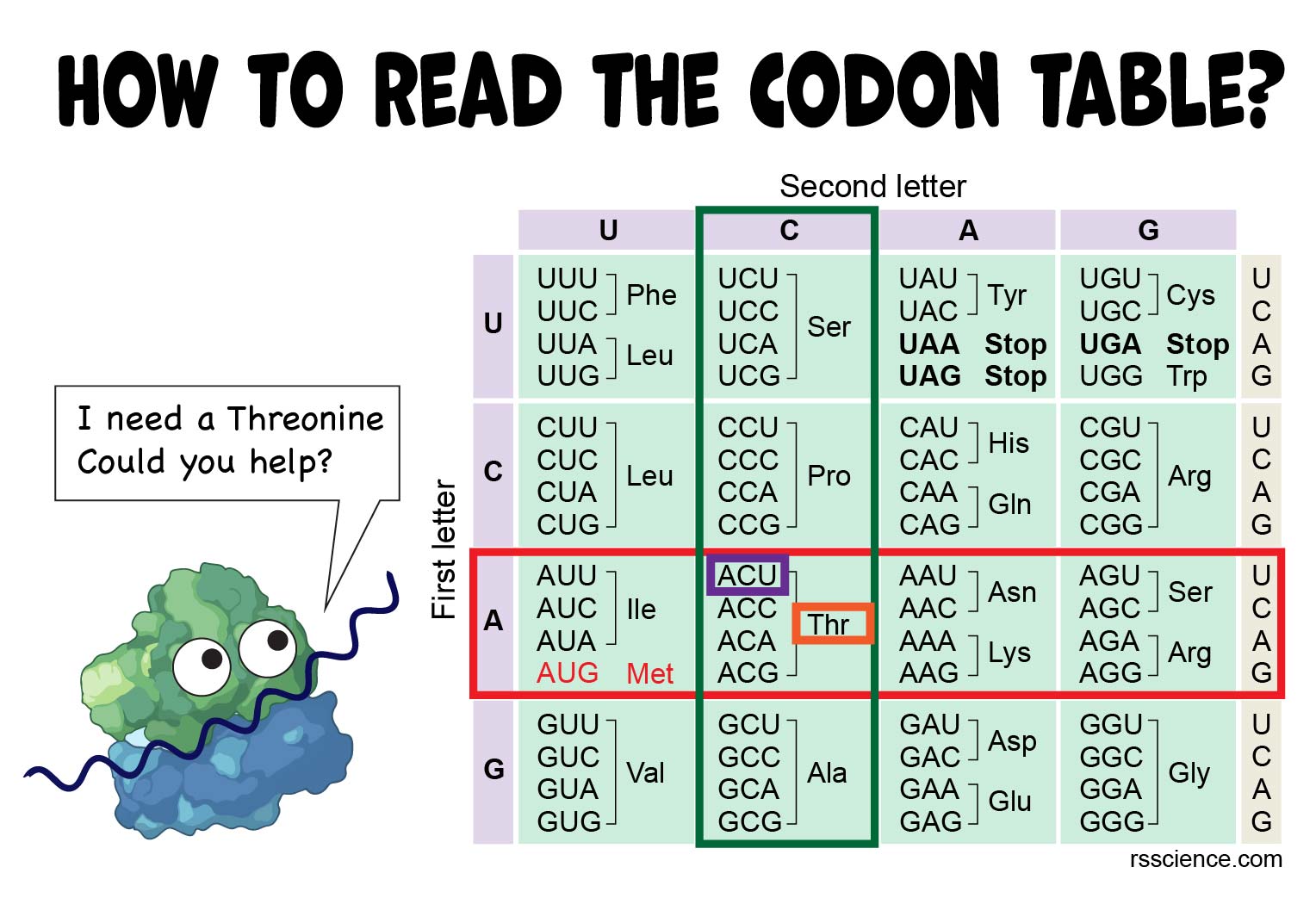
How To Use The Codon Chart YouTube
Web use this option to calculate the codon adaptation index (cai) from a protein alignment that has been translated to a dna alignment using (a) a unique codon usage table as a. Web the codon adaptation index (cai) developed by sharp and li [ 4 ], rapidly became one of the most used indices. Web.
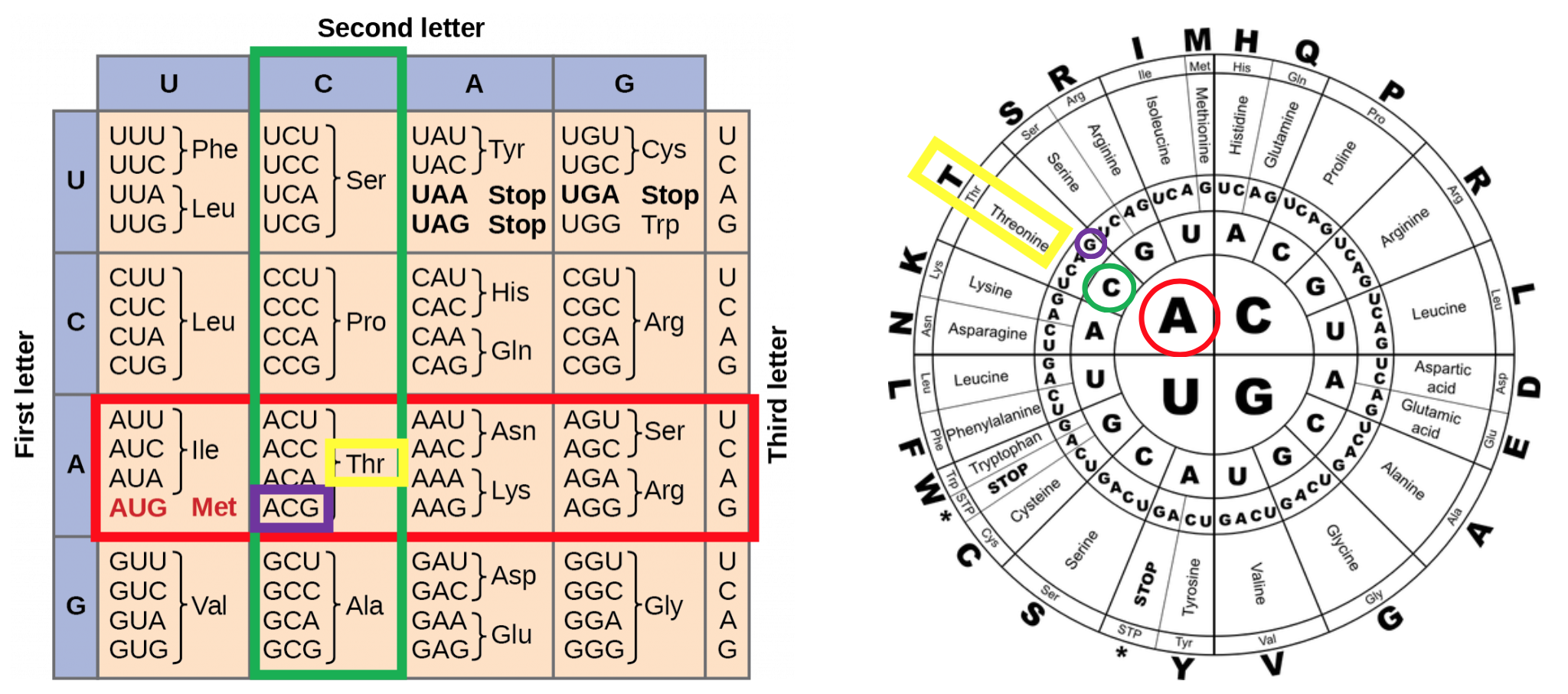
Amino Acid Codon Wheel SigmaAldrich
Web important considerations when selecting a codon usage calculator. Web use this option to calculate the codon adaptation index (cai) from a protein alignment that has been translated to a dna alignment using (a) a unique codon usage table as a. Codon usage calculator returns the number and frequency of each codon type and associated..
Codon Usage Calculator Web crucially, for analyzing codon usage in common host organisms %minmax requires only the coding sequence as input; This online codon usage calculator accept one raw sequence and calculate the outputed codon usage table according to the genetic code. Web the gcua tool displays the codon quality either in codon usage frequency values or relative adaptiveness values. Web this codon optimization tool is integrated with our complexity checker and api sci tools for a seamless design and ordering experience. Web the codon usage tables are linked to a taxonomy tree to allow comparative analysis of the codon usage frequencies.
Web Resources » Bioinformatics Tools ** This Online Tool Shows Commonly Used Genetic Codon Frequency Table In Expression Host Organisms Including Escherichia Coli And Other.
Web accurately quantifying codon usage bias for different organisms is useful not only for codon optimization, but also for evolutionary and translation studies:. Usage table submit a dna sequence in. This online codon usage calculator accept one raw sequence and calculate the outputed codon usage table according to the genetic code. Enter your dna/rna sequence (s) in the box below in fasta or plaintext format:
This Online Tool Shows Codon Usage Frequency Table For Some Protein Expression Systems, Including E.coli, Yeast ( Pichia Pastoris Or.
Search the database for the codon usage table of your organism of interest. Web %minmax calculator gene sequence (help): Web codon usage advertisement codon usage genetic code: The rapid growth of interest in the effects of synonymous codon usage on gene expression.
Cordon Provides Tools For Analysis Of Codone Usage (Cu) In Various Unannotated Or Kegg/Cog Annotated Dna Sequences.
Web this online tool calculates cai according to the relative synonymous codon usage of a reference sequence or existing expression host organisms. Perform random reverse translation, as control (slower) (help) please use the. Calculator accepts fasta dna sequence. Paste the sequences either as raw data ( iupac code) or as one or more fasta sequences.
Web The Codon Adaptation Index (Cai) Developed By Sharp And Li [ 4 ], Rapidly Became One Of The Most Used Indices.
Web crucially, for analyzing codon usage in common host organisms %minmax requires only the coding sequence as input; Web codon usage codon usage accepts one or more dna sequences and returns the number and frequency of each codon type. Web important considerations when selecting a codon usage calculator. The cai is a measure of the synonymous codon.